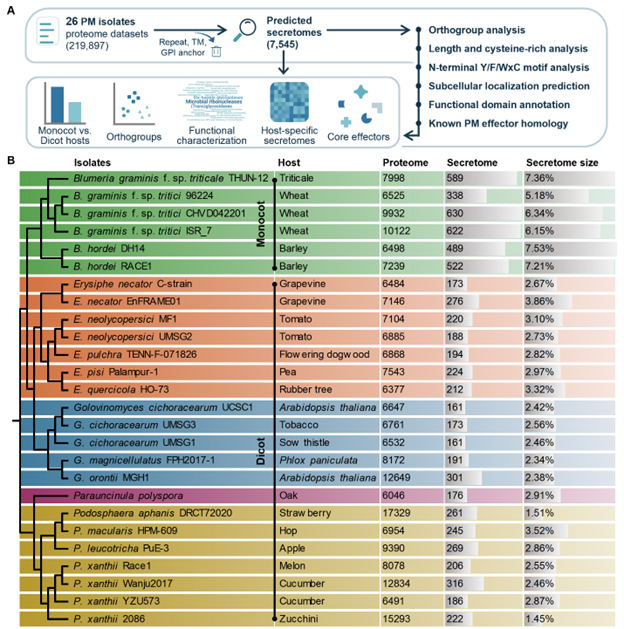
PM3D (Powdery Mildew Multi-Omics Data Database) provides comprehensive proteome, secretome, effector, and functional annotation data for 26 races of powdery mildew fungi. Integrating functional predictions from SignalP, DeepTMHMM, InterProScan, and AlphaFold2, PM3D enables researchers to explore fungal proteins, their Functional annotation Characterization and 3D structures for plant–pathogen interaction studies.
Data Analysis Pipeline Integrated with PM3D:
- SignalP0 Identifies proteins with signal peptides, marking them as potentially secreted.
- DeepTMHMM Predicts transmembrane helices, helping distinguish membrane-bound proteins from soluble secreted proteins.
- InterProScan Functional domain and motif annotation across the proteome to identify enzymatic, structural, or effector-like domains.
- 0 Integrates results from signal peptide and transmembrane analysis to determine the Effectors candidates, i.e., proteins likely secreted by the fungus to interact with the host.
- Structural Prediction (AlphaFold2) Provides predicted 3D protein structures of secreted and effector proteins, enabling structural and functional characterization.
Database Purpose:
PM3D is designed to serve researchers studying plant–pathogen interactions, offering:
- Access to curated proteomes of 26 powdery mildew races
- Annotated secretomes and effector candidates
- Functional domain and localization predictions
- Structural models for downstream analysis and hypothesis generation
PIPELINE work flow: